
Sign in to participate in a fun event
Write your own genetic module
Past
Thu, Oct 31, 2024
12:00 - 14:00 GMT+1
Halle Moos
Moosdorfstraße 7-9, 12435 Berlin, Germany
Copy Address
Venue Detail
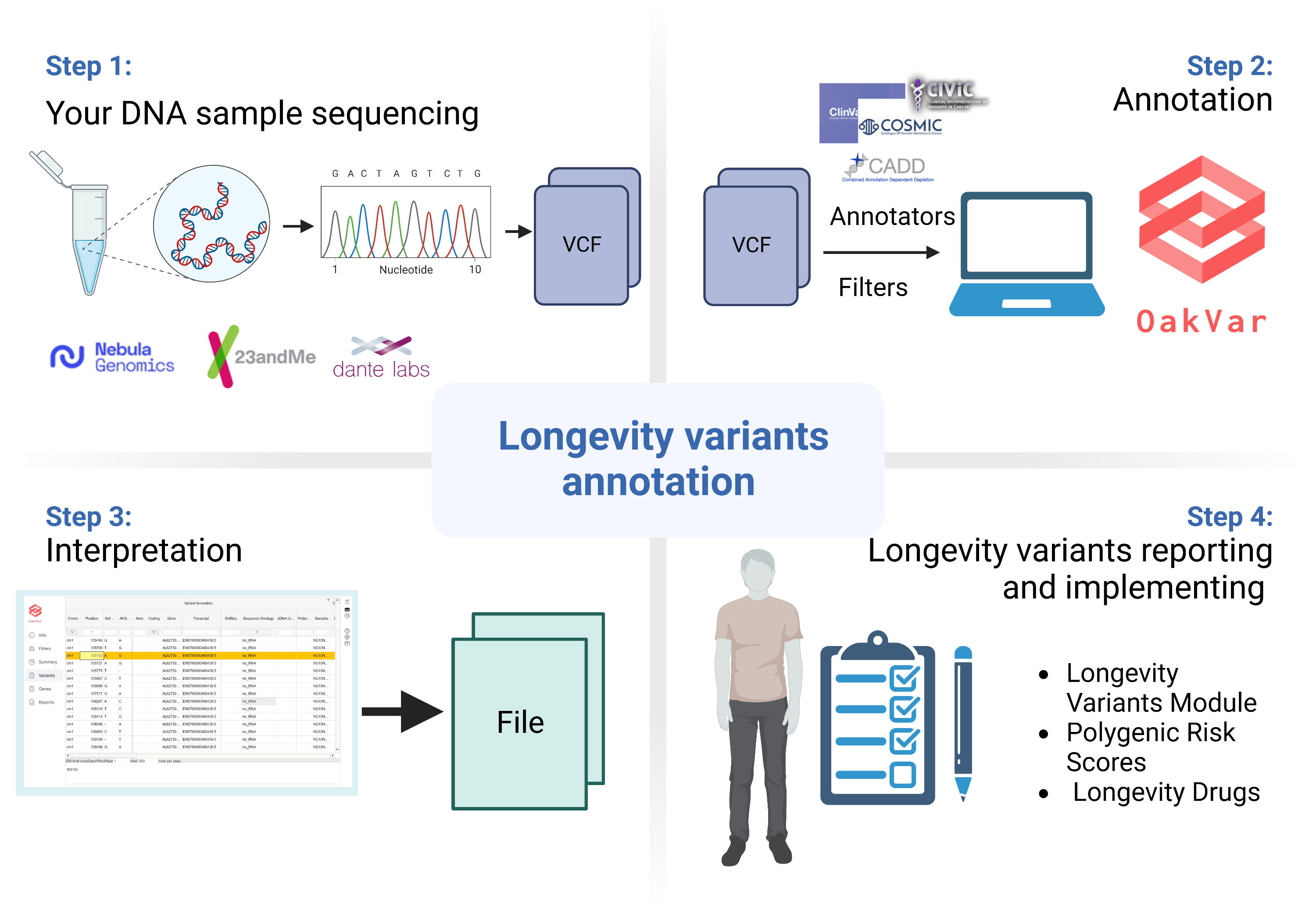
We will write our own genomic modules so you can annotate genomes with them to make genomic reports
To participate on the coding side you must setup oakvar and dependencies The instructions are at https://github.com/dna-seq/quick-dna
It is overnight download because of huge dbsnp
Also, think which genetic variants do you want to filter or annotate with the module. If you don't have coding skills you can participate from biology side by finding genetic variants of interest.
So far ideas:
pathogenic filters
intelligence
mirochodria
Note, stuff that we already have is: https://dna-seq.github.io/index.html#modules
Comments